Current preprints
Abasi LS, Elathram N, Movva M, Deep A, Corbett KD, Debelouchina GT* (2023). Phosphorylation regulates tau's phase separation behavior and interactions with chromatin bioRxiv, preprint. DOI: httpss://doi.org/10.1101/2023.12.21.572911
In this manuscript, we focus on the interactions between the Alzheimer's disease related protein tau and chromatin. While tau is primarily known as a microtubule binding protein that works in the cytoplasm, there is evidence that tau is also found in heterochromatin domains in the nucleus. In this in vitro study, we investigate tau's phase separation propensity and interactions with DNA, reconstituted nucleosome arrays, and heterochromatin proteins. We show that these properties are dependent on tau's phosphorylation state. This study interrogates a possible disease-state mechanism for tau-mediated heterochromatin disruption that is distinct from the amyloid formation hypothesis.
In this manuscript, we focus on the interactions between the Alzheimer's disease related protein tau and chromatin. While tau is primarily known as a microtubule binding protein that works in the cytoplasm, there is evidence that tau is also found in heterochromatin domains in the nucleus. In this in vitro study, we investigate tau's phase separation propensity and interactions with DNA, reconstituted nucleosome arrays, and heterochromatin proteins. We show that these properties are dependent on tau's phosphorylation state. This study interrogates a possible disease-state mechanism for tau-mediated heterochromatin disruption that is distinct from the amyloid formation hypothesis.
Yoon J, Zhang YM, Her C, Grant RA, Ponomarenko AM, Ackermann BE, Debelouchina GT, Shoulders MD* (2023). The Immune-Evasive Proline 283 Substitution in Influenza Nucleoprotein Increases Aggregation Propensity Without Altering the Native Structure bioRxiv, preprint. DOI: httpss://doi.org/10.1101/2023.09.08.556894
In collaboration with the Shoulders Lab (MIT), we use solution NMR spectroscopy to probe the effects of a single point mutation on the structure and dynamics of the influenza nucleoprotein.
In collaboration with the Shoulders Lab (MIT), we use solution NMR spectroscopy to probe the effects of a single point mutation on the structure and dynamics of the influenza nucleoprotein.
Phan TM, Kim YC, Debelouchina GT*, Mittal J* (2023). Interplay between charge distribution and DNA in shaping HP1 paralog phase separation and localization bioRxiv, preprint. DOI: httpss://doi.org/10.1101/2023.05.28.542535
In collaboration with the Mittal group (Texas A&M), we decode how the sequence differences between the three human HP1 paralogs underlie differences in their phase separation behavior, potentially underlying differences in function and localization in the nucleus.
In collaboration with the Mittal group (Texas A&M), we decode how the sequence differences between the three human HP1 paralogs underlie differences in their phase separation behavior, potentially underlying differences in function and localization in the nucleus.
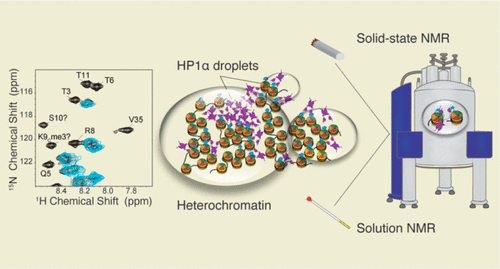
2023Elathram N, Ackermann BE, Clark ET, Dunn SR, Debelouchina GT* (2023). Phosphorylated HP1α-Nucleosome Interactions in Phase Separated Environments Journal of the American Chemical Society, 145, 44, 23994-24004
DOI: httpss://doi.org/10.1021/jacs.3c06481 We use solution and solid-state NMR spectroscopy to detail the scope of interactions between phosphorylated HP1α and the nucleosome, a landscape with many transient interactions that are difficult to detect by most biophysical techniques. |
|
Kent JE, Ackermann BE, Debelouchina GT*, Marassi FM* (2023). Dynamic Nuclear Polarization Illuminates Key Protein-Lipid Interactions in the Native Bacterial Cell Envelope Biochemistry, 62, 15, 2252-2256. DOI: httpss://doi.org/10.1021/acs.biochem.3c00262
In collaboration with the Marassi Lab (Medical College of Wisconsin), we harness DNP-enhanced solid-state NMR spectroscopy to study the interactions of the plague bacterium protein Ail with the surrounding native cell membrane. For the first time, DNP enabled us to detect elusive key contacts between the outer loop arginine residues on the protein and the lipopolysaccharide layer of the cell envelope. These interactions support a model where the Ail protein potentially remodels the bacterium cell envelope environment as part of its mechanism of action and host invasion. |
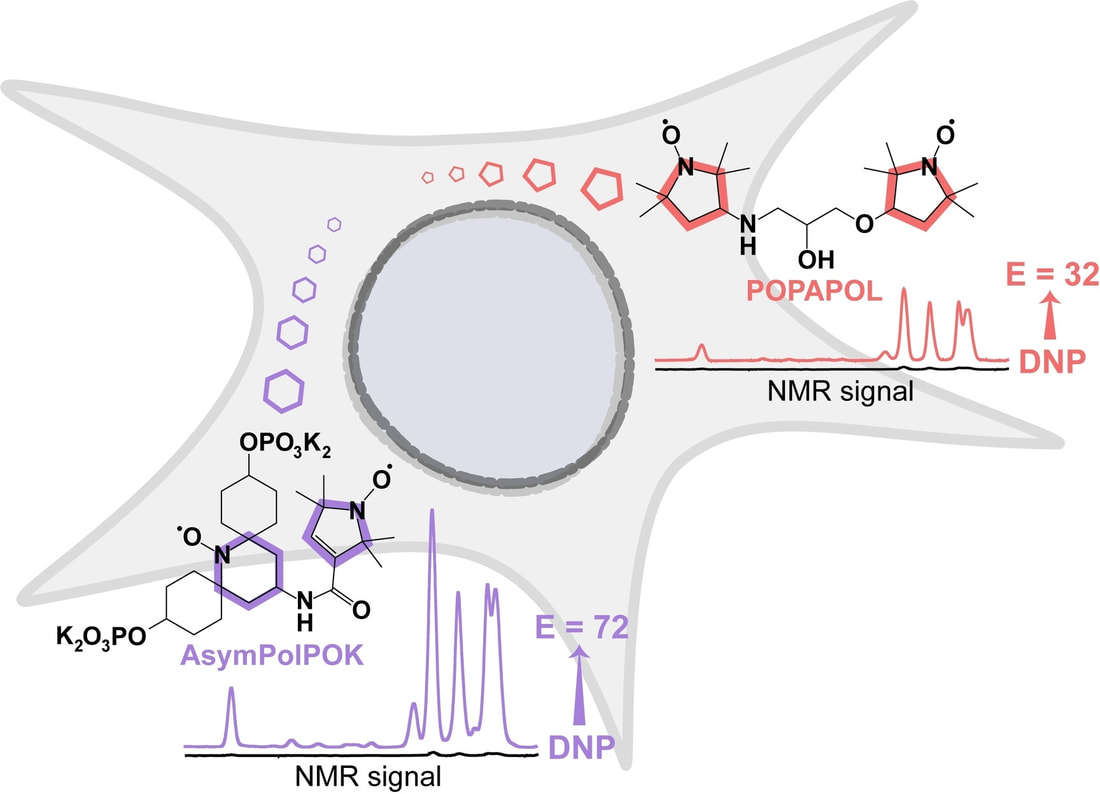
2022Ackermann BE, Lim BJ, Elathram N, Narayanan S, Debelouchina GT* (2022). A Comparative Study of Nitroxide-Based Radicals for Dynamic Nuclear Polarization in Cellular Environments ChemBioChem, 23, e202200577. DOI: httpss://doi.org/10.1002/cbic.202200577
We investigate the stability of commonly used DNP polarization agents in the reducing cellular environment and show that increased steric hindrance around the nitroxide-based radical can improve DNP performance in this setting. This information is important to design more efficient DNP polarization agents for sensitivity-enhanced NMR spectroscopy in cells. |
|
Her C#, Phan TM#, Jovic N, Kapoor U, Ackermann BE, Rizuan A, Kim Y, Mittal J* & Debelouchina GT* (2022). Molecular interactions underlying the phase separation of HP1α: Role of phosphorylation, ligand and nucleic acid binding Nucleic Acids Research, 50 (22), 12702-12722. DOI: httpss://doi.org/10.1093/nar/gkac1194
# Authors contributed equally We couple in vitro phase separation assays with the computational approaches of the Mittal group (Texas A&M) to understand the interactions that drive the formation and modulation of HP1α liquid droplets. HP1α is a key participant in the formation of gene silencing domains in the nucleus. |
|
Berkeley RF & Debelouchina GT* (2022). Chemical tools for study and modulation of biomolecular phase transitions. Chemical Science, 13. 14226-14245. DOI:
httpss://doi.org/10.1039/D2SC04907D A perspective on state-of-the-art tools to study intrinsically disordered proteins (IDPs) in the context of liquid-liquid phase separation (LLPS). For example, we discuss site-specific modifications for biophysical characterization and small molecule modulators of LLPS propensity. |
|
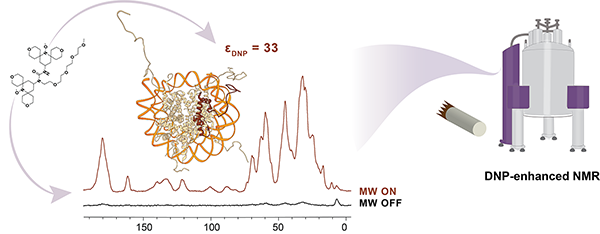
Elathram N, Ackermann BE & Debelouchina GT* (2022). DNP-enhanced solid-state NMR of chromatin polymers Journal of Magnetic Resonance Open, 10-11, 100057. DOI: httpss://doi.org/10.1016/j.jmro.2022.100057
We premiere the use of DNP solid-state NMR spectroscopy for structural biology studies of chromatin. The sensitivity enhancements afforded by DNP allow the detection of protein-DNA contacts of much smaller amounts of precious reconstituted chromatin samples. |
|
Clark ET#, Sievers EE# & Debelouchina GT* (2022). A chemical biology primer for NMR spectroscopists Journal of Magnetic Resonance Open, 10-11, 100044. DOI: httpss://doi.org/10.1016/j.jmro.2022.100044
#Authors contributed equally A tutorial for engineering protein constructs suitable for NMR spectroscopy. We cover techniques such as segmental isotope labeling, unnatural amino acid incorporation, post-translational modifications, probe conjugation, and more! |
Podolsky KA, Masubuchi T, Debelouchina GT, Hui E* & Devaraj NK* (2022). In Situ Assembly of Transmembrane Proteins from Expressed and Synthetic Components in Giant Unilamellar Vesicles ACS Chemical Biology, 17 (5), 1015-1021. DOI: httpss://doi.org/10.1021/acschembio.2c00013
In collaboration with the Hui and Devaraj labs at UCSD, we demonstrate how inteins can be used to build transmembrane proteins in lipid vesicles.
In collaboration with the Hui and Devaraj labs at UCSD, we demonstrate how inteins can be used to build transmembrane proteins in lipid vesicles.
2021Ackermann BE & Debelouchina GT* (2021). Emerging contributions of solid-state NMR spectroscopy to chromatin structural biology Frontiers in Molecular Biosciences, 8, 741581. DOI: httpss://doi.org/10.3389/fmolb.2021.741581
This review summarizes the recent literature where solid-state NMR spectroscopy was used to investigate both the rigid and dynamic components of nucleosomes and nucleosome arrays. We also review the current strategies for chromatin sample preparation and offer insights into how chemical biology tools can contribute to future structural studies of chromatin and chromatin interacting proteins. |
|
Berkeley RF, Kashefi M & Debelouchina GT* (2021). Real-time observation of structure and dynamics during the liquid-to-solid transition of FUS LC. Biophysical Journal, 120 (7), 1276. DOI: httpss://doi.org/10.1016/j.bpj.2021.02.008
We track the liquid droplet to fibril transition process of the low complexity domain of the RNA-binding protein FUS. Using solid-state NMR, we show that this process results in β-sheet structures that appear distinct from fibrils grown in the absence of liquid droplets. We also use coarse-grained molecular dynamics simulations to show how a disease-relevant mutation leads to more rapid fibril formation. |
2020Lim BJ, Berkeley RF & Debelouchina GT* (2020). Fused split inteins: Tools for introducing multiple protein modifications. In: Vila-Perelló M. (eds) Expressed Protein Ligation. Methods in Molecular Biology, vol 2133. Humana, New York, NY. DOI: httpss://doi.org/10.1007/978-1-0716-0434-2_8
A protocol highlighting the use of inteins as a tool for tagless protein purification without the need to use proteases. This methodology also enables the installation and purification of multiple protein modifications and illustrates the growing versatility of inteins as a protein engineering tool. |
|
Lim BJ#, Ackermann BE# & Debelouchina GT* (2020). Targetable tetrazine-based dynamic nuclear polarization agents for biological systems. ChemBioChem 21, 1315. DOI: httpss://doi.org/10.1002/cbic.201900609
#Authors contributed equally We present a method for the selective targeting of dynamic nuclear polarization (DNP) agents to proteins in complex environments such as cellular lysates. Our strategy offers spatially localized NMR signal enhancement and opens the door to new avenues of in-cell NMR and DNP polarization transfer studies. |

|
2019Ackermann BE & Debelouchina GT* (2019). Heterochromatin protein HP1α gelation dynamics revealed by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. 131 (19), 6366.
DOI: httpss://doi.org/10.1002/anie.201901141 We showcase solid-state NMR spectroscopy as a powerful method for illuminating the structure and dynamics of proteins undergoing liquid-liquid phase separation. Here, we follow a time course of the nuclear protein HP1α as it undergoes a transition from liquid droplets to a gel state, and identify residues that mediate this transition. The gelation process is slowed by the presence of nucleosome arrays, suggesting that chromatin can also modulate the material state of its surrounding environment. |
Publications prior to UCSD:
Beh LY, Debelouchina GT, Clay DM, Thompson RE, Lindblad KA, Hutton ER, Bracht JR, Sebra RP, Muir TW. and Landweber LF (2019). Identification of a DNA N6-Adenine Methyltransferase Complex and Its Impact on Chromatin Organization. Cell 177 (7), 1781.
Iadanza MG, Silvers R, Boardman J, Smith HI, Karamanos TK, Debelouchina GT, Su Yongchao, Griffin RG, Ranson NA and Radford SE (2018). The structure of a β2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat. Comm. 9 (1), 4517.
Wojcik F, Dann GP, Beh LY, Debelouchina GT, Hofmann R and Muir TW (2018). Functional crosstalk between histone H2B ubiquitylation and H2A modifications and variants. Nat. Comm. 9 (1), 1394.
Debelouchina GT and Muir TW (2017). A Molecular Engineering Toolbox for the Structural Biologist.
Q. Rev. Biophys. 50, e7.
Debelouchina GT, Gerecht K, Muir TW (2017). Ubiquitin utilizes an acidic surface patch to alter chromatin structure.
Nat. Chem. Biol. 13, 105-110.
Frederick KK, Michaelis VK, Caporini MA, Andreas LB, Debelouchina GT, Griffin RG, Lindquist S. (2017). Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register.
Proc. Natl. Acad. Sci. U.S.A. 114, 3642-3647.
Johnson JG, Wang B, Debelouchina GT, Novick RP, Muir TW (2015). Increasing AIP Macrocycle Size Reveals Key Features of agr Activation in Staphylococcus aureus. Chembiochem 16, 1093-100.
Liu Z, Frutos S, Bick MJ, Vila-Perello M, Debelouchina GT, Darst SA, Muir TW (2014). Structure of the Branched Intermediate in Protein Splicing. Proc. Natl. Acad. Sci. U.S.A. 111, 8422-7.
Su Y, Sarell CJ, Eddy ET, Debelouchina GT, Andreas LB, Pashley CL, Radford SE, Griffin RG (2014). Secondary Structure in the Core of Amyloid Fibrils Formed from Human β2m and Its Truncated Variant ΔN6. J. Am. Chem. Soc. 136, 6313-25.
Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S (2014). Distinct Prion Strains Are Defined by Amyloid Core Structure and Chaperone Binding Site Dynamics. Chem. Biol. 21, 295-305.
Debelouchina GT, Bayro MJ, Fitzpatrick AWP, Ladizhansky V, Colvin MT, Caporini MA, Jaroniec CP, Bajaj VS, Rosay M, MacPhee C, Vendruscolo M, Maas WE, Dobson CM, Griffin RG (2013). Higher Order Amyloid Fibril Structure by MAS NMR and DNP Spectroscopy. J. Am. Chem. Soc. 135, 19237-47.
Fitzpatrick AWP, Debelouchina GT, Bayro MJ, Clare DK, Caporini MA, Bajaj VS, Jaroniec CP, Wang LC, Ladizhansky V, Muller SA, MacPhee CE, Waudby CA, Mott HR, De Simone A, Knowles TPJ, Saibil HR, Vendruscolo M, Orlova EV, Griffin RG, Dobson CM (2013). Atomic Structure and Hierarchical Assembly of a Cross-β Amyloid Fibril.
Proc. Natl. Acad. Sci. U.S.A. 110, 5468-73.
Sarell CJ, Woods LA, Su YC, Debelouchina GT, Ashcroft AE, Griffin RG, Stockley PG, Radford SE (2013). Expanding the Repertoire of Amyloid Polymorphs by Co-polymerization of Related Protein Precursors. J. Biol. Chem. 288, 7327-37.
Bayro MJ, Debelouchina GT, Eddy MT, Birkett, NR, Rosay M, Maas WE, Dobson CM, Griffin RG (2011). Intermolecular Structure Determination of Amyloid Fibrils by Magic-Angle Spinning NMR Spectroscopy and Dynamic Nuclear Polarization. J. Am. Chem. Soc. 133, 13967-74.
Hu KN, Debelouchina GT, Smith AA, Griffin RG (2011). Quantum Mechanical Theory of Dynamic Nuclear Polarization in Solid Dielectrics. J. Chem. Phys. 134, 125105.
Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG (2010). Intermolecular Alignment in β2-Microglobulin Amyloid Fibrils. J. Am. Chem. Soc. 132, 17077-9.
Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG (2010). Magic Angle Spinning NMR Analysis of β2-Microglobulin Amyloid Fibrils in Two Distinct Morphologies. J. Am. Chem. Soc. 132, 10414-23.
Debelouchina GT*, Bayro MJ*, van der Wel PCA, Caporini MA, Barnes AB, Rosay M, Maas WE, Griffin (2010). Dynamic Nuclear Polarization Enhanced Solid-State NMR Spectroscopy of GNNQQNY Crystals and Amyloid Fibrils.
Phys. Chem. Chem. Phys. 12, 5911-9.
* Authors contributed equally
Flook MM, Gerber LCH, Debelouchina GT, Schrock R (2010). Z-Selective and Syndioselective Ring-Opening Metathesis Polymerization (ROMP) Initiated by Monoaryloxidepyrrolide (MAP) Catalysts. Macromolecules 43, 7515-7522.
Dane EL, Maly T, Debelouchina GT, Griffin RG, Swager TM (2009). Synthesis of a BDPA-TEMPO Biradical.
Org. Lett. 11, 1871-4.
Maly T, Debelouchina GT, Bajaj VS, Hu KN, Joo CG, Mak-Jurkauskas ML, Sirigiri JR, van der Wel PCA, Herzfeld J, Temkin RJ, Griffin RG (2008). Dynamic Nuclear Polarization at High Magnetic Fields. J. Chem. Phys. 128, 052211.
Galia Debelouchina - University of California, San Diego - Department of Chemistry and Biochemistry - © Debelouchina Lab 2024